

| LTPDA Toolbox™ | contents |   |
We can, of course, do the same injections but with the full LPF model and with noise switched on.
In Topic 2 you already saw how to simulate an LPF model with all the noises switched on. If we combine that with the signal injections we've been doing in this topic, then a simulation with noise and injected signals can be done like this:
% Create a standard LPF model
lpf = ssm(plist('built-in', 'LPF'));
% Generate suitable covariance matrix for all inputs
cov = lpf.generateCovariance;
% Create some time-series analysis object to inject
aSignal = ao(plist('tsfcn', '1e-7*sin(2*pi*0.005*t)', 'fs', 1/lpf.timestep, 'nsecs', 10000));
% Create the plist to configure simulate
sim_pl = plist('AOS', aSignal, 'AOS Variable Names', 'GUIDANCE.ifo_x1', ...
'return outputs', {'DELAY_IFO.x1', 'DELAY_IFO.x12'}, ...
'CPSD variable names', cov.find('names'), ...
'CPSD', cov.find('cov'));
% Run the simulation
out = simulate(lpf, sim_pl);
% Unpack the outputs
[o1, o12] = unpack(out);
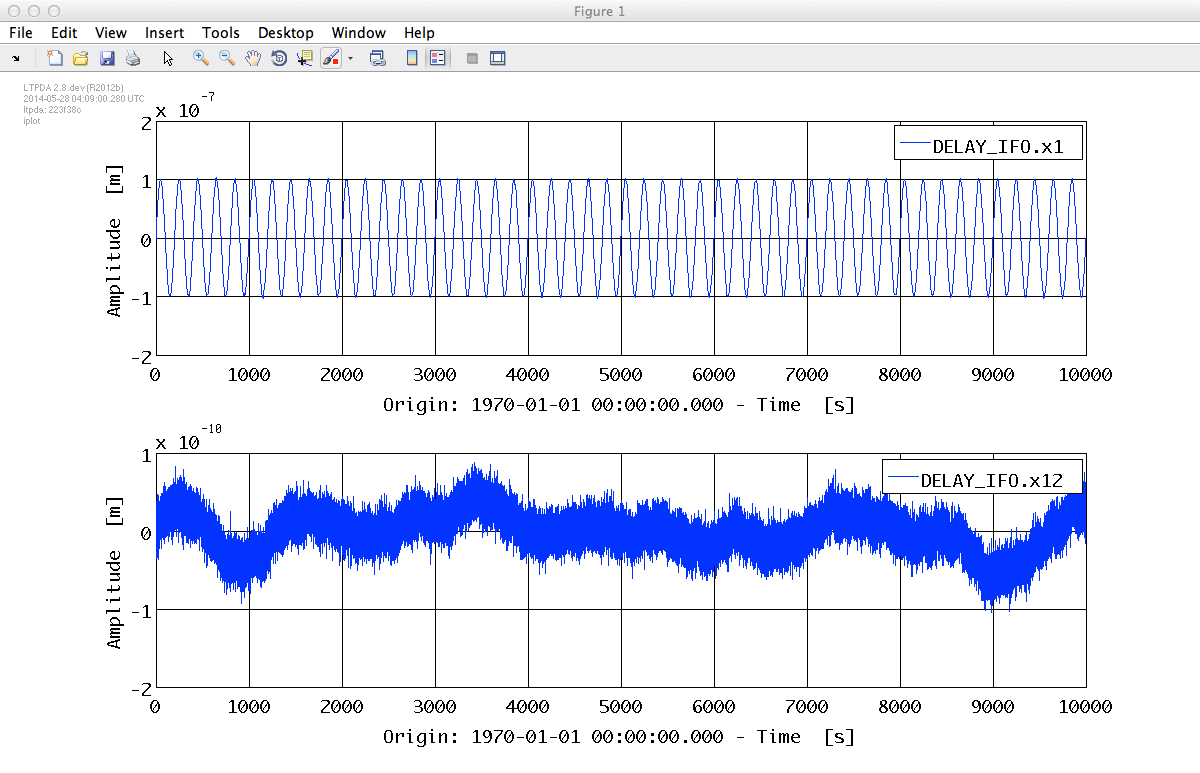
% Plot on subplots
plot_pl = plist('arrangement', 'subplots');
iplot(o1, o12, plot_pl);
% Create a PSD plist with 16 averages, linear segment-wise detrending
psd_pl = plist('navs', 16, 'order', 1, 'win', 'BH92');
% Estimate PSDs
[o1_xx, o12_xx] = psd(o1, o12, psd_pl);
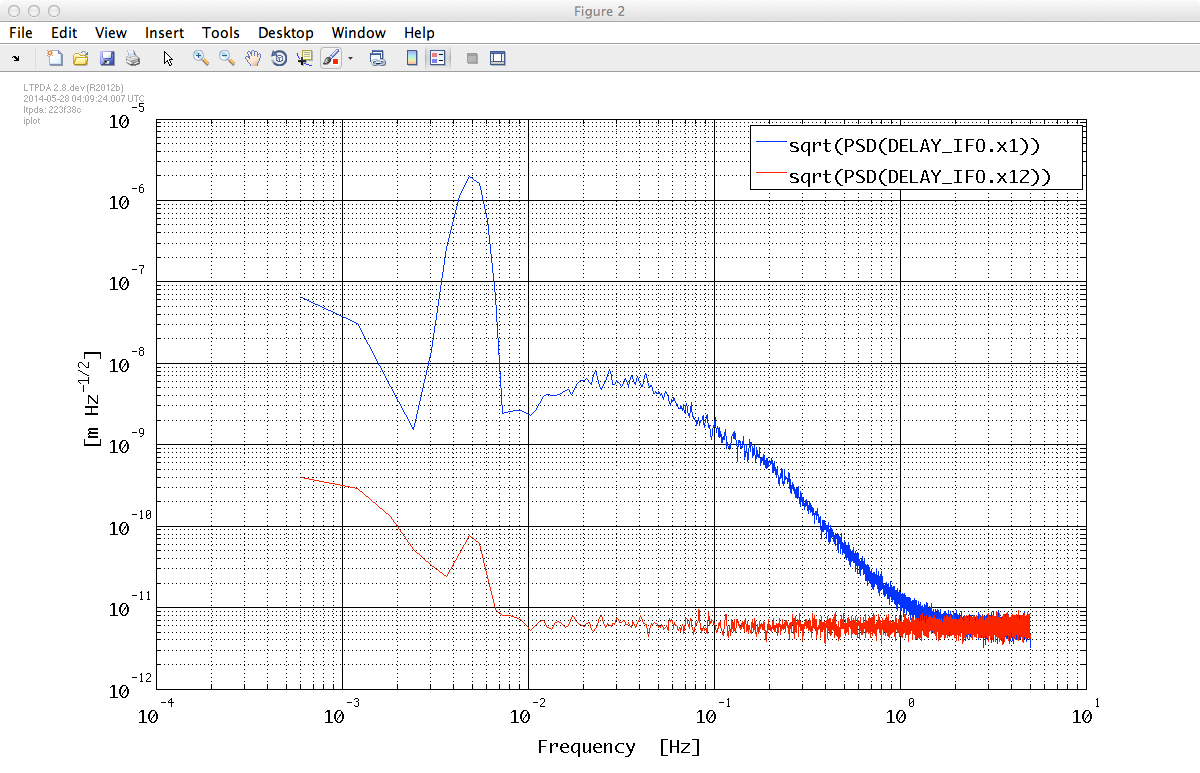
% Plot the ASDs
iplot(sqrt(o1_xx), sqrt(o12_xx));
 |
Estimate tranfser functions from simulated signals, compare with Bode estimates | Topic 5 - Introduction to system identification of LPF. |  |
©LTP Team